Development of an analytical method to estimate gene expression by cell-cell interaction
CCPLS reveals cell-type-specific spatial dependence of transcriptomes in single cells.
Our bodies are composed of a wide variety of cells, which interact with each other to ensure that developmental processes proceed properly and that homeostasis is maintained in tissues and organs. When this cell-cell interaction is disrupted, it can lead to disease.
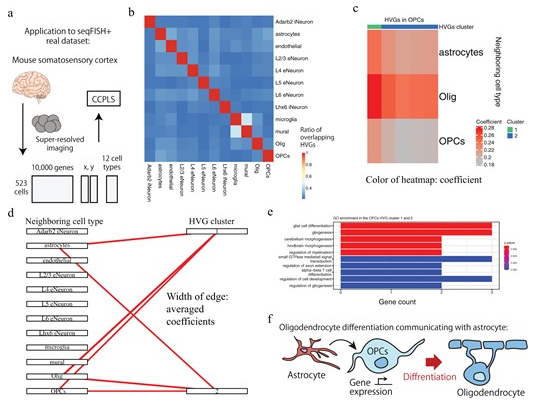
In this study, we developed CCPLS, an information analysis method that uses one-cell spatial transcriptome data, which simultaneously measures cell function and spatial coordinates, to estimate the effects of neighboring cells on each other's gene expression. This method estimates the relationship between the types of neighboring cell types and the resulting gene expression when focusing on a certain cell type. Evaluation experiments using simulated data have shown that CCPLS can estimate cell-cell interactions with high accuracy. In addition, specific cell-cell interactions could be extracted from examples of application to real data in the brain and colon, demonstrating the effectiveness of this method.
→
Journal Publication - Bioinformatics 【DOI】 10.1093/bioinformatics/btac599
CCPLS reveals cell-type-specific spatial dependence of transcriptomes in single cells.
→ Research Outline (PDF In Japanese language)