Molecular signals within excitatory neurons regulate sleep
Elucidating the mechanism that determines the quantity and quality of sleep.
Sleep is necessary for everyone, but why sleep is needed at all is a mystery to this day. The research team focused on an enzyme (SIK3) which is key to solving this mystery. This enzyme regulates the chain of reactions in the brain involved in sleep, forming a molecular signal which regulates the quality and quantity of sleep. However, it was not known what kind of molecules and chains of molecules SIK3 forms for sleep regulation, or through which cells it is determined. In this study, the details of the chain (molecular signal) and the genes regulated by this molecular signal were clarified for the first time ever internationally.
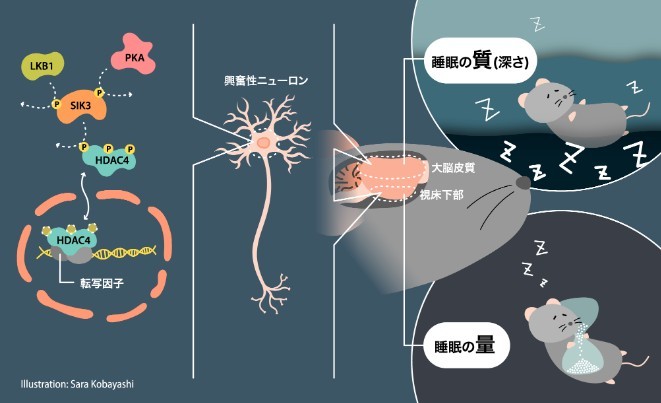
Sleep is essential for physical and mental health. Sleep disorders increase the risk of mental disorders, diabetes, heart disease, and dementia such as Alzheimer's disease, and decrease brain performance during the day. It is said that many people in Japan have sleep debt (mental and physical disorders associated with sleep deprivation). Through understanding the mechanisms that control the quantity and quality of sleep, it is expected to contribute to the development of new methods of sleep control and treatments for sleep disorders.
→
Journal Publication A - Nature 【DOI】 10.1038/s41586-022-05450-1
Kinase signalling in excitatory neurons regulates sleep quantity and depth.
→
Journal Publication B - Nature 【DOI】 10.1038/s41586-022-05510-6
A signaling pathway for transcriptional regulation of sleep amount in mice.
→ Research Outline (PDF In Japanese language)
2022 Chemo-Sero Therapeutic Research Institute Grant

Professor Mamiko Sakata-Yanagimoto (坂田-柳元 麻実子), who heads up the Advanced Hemato Oncology Laboratory in the TMRC Integrated Research Division, was selected for the Japan 2022 Chemo-Sero Therapeutic Research Institute Grant. The funding will go to support her lab's important research elucidating the pathogenesis of age pre-related malignant lymphoma.
→ "Kaketsuken" Press Release (PDF In Japanese language)
→
TMRC Integrated Research Division - Advanced Hemato Oncology Lab
• Quiescent muscle stem cells express the adhesion G-protein-coupled receptor Gpr116
• Gpr116-deficient muscle stem cells are incapable of maintaining quiescence
• The GPR116 Stachel agonist peptide prevents MuSC activation
• GPR116 signaling is mediated in part by nuclear activity of β-arrestin1
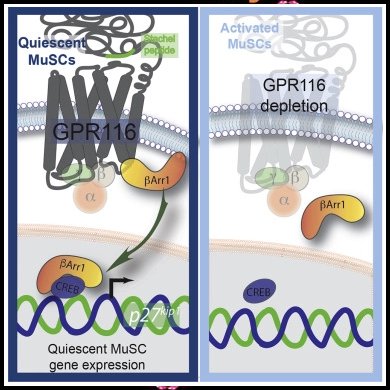
Skeletal muscle is populated with a reservoir of quiescent muscle stem cells (MuSCs), which regenerate the tissue after injury. Here, we show that the adhesion G-protein-coupled receptor Gpr116 is essential for long-term maintenance of the MuSC pool. Quiescent MuSCs express high levels of Gpr116, which is rapidly downregulated upon MuSC activation. MuSCs deficient for Gpr116 exhibit progressive depletion over time and are defective in self-renewal. Adhesion G-protein-coupled receptors contain an agonistic peptide sequence, called the “Stachel” sequence, within their long N-terminal ectodomains. Stimulation of MuSCs with the GPR116 Stachel peptide delays MuSC activation and differentiation. Stachel peptide stimulation of GPR116 leads to strong interaction with β-arrestins. Stimulation of GPR116 increases the nuclear localization of β-arrestin1, where it interacts with cAMP response element binding protein to regulate gene expression. Altogether, we propose a model by which GPR116 maintains the MuSC pool via nuclear functions of β-arrestin1.
→
Cell Reports Volume 41, ISSUE 7, 111645, November 15, 2022
https://doi.org/10.1016/j.celrep.2022.111645
→ Research Outline (PDF In Japanese language)
Digital Medicine Strategy Division - Fourth Seminar Session
Transborder Medical Research Center
Digital Medicine Strategy Division
Online Zoom Event,
November 30th, 2022 (Wednesday),
18:00 - 19:00
(Please Note: Seminar will be presented in Japanese language only)

→
Professor Sakata-Yanagimoto received Japan Cancer Association (JCA) Mauvernay Award.
Professor Sakata-Yanagimoto of TMRC and Department of Hematology at University of Tsukuba, received the 2022 Japan Cancer Association (JCA) Mauvernay Award within the “Translational Research” catergory. Professor Sakata-Yanagimoto was recognized for her exceptional research related to “Translational research targeting intractable lymphomas.”
The Japanese Cancer Association and Debiopharm jointly established the JCA-Mauvernay Award to recognize the cutting edge oncology research and to encourage Japanese researchers to make their work available to other parts of the world.”
Read More → (from Japan Cancer Association (JCA) - External Website)
Award Information → (Japan Cancer Association (JCA) - External Website)

“The “Basic Project for Supporting Life Science and Drug Discovery Research” is a project based on a wide range of life science-related research in Japan, with the aim of linking outstanding research results to practical research and development, such as drug discovery research. ” Read More → (BINDS Website - Available in Japanese Language Only)
Development of an analytical method to estimate gene expression by cell-cell interaction
CCPLS reveals cell-type-specific spatial dependence of transcriptomes in single cells.
Our bodies are composed of a wide variety of cells, which interact with each other to ensure that developmental processes proceed properly and that homeostasis is maintained in tissues and organs. When this cell-cell interaction is disrupted, it can lead to disease.
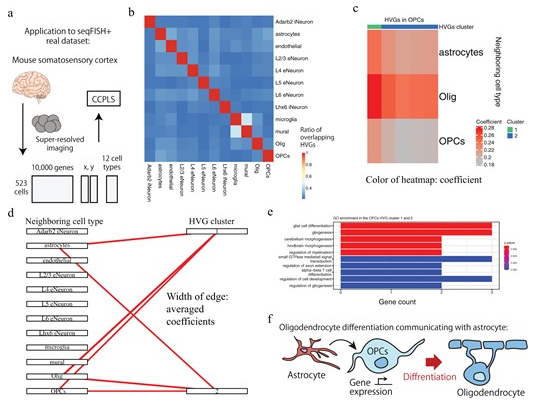
In this study, we developed CCPLS, an information analysis method that uses one-cell spatial transcriptome data, which simultaneously measures cell function and spatial coordinates, to estimate the effects of neighboring cells on each other's gene expression. This method estimates the relationship between the types of neighboring cell types and the resulting gene expression when focusing on a certain cell type. Evaluation experiments using simulated data have shown that CCPLS can estimate cell-cell interactions with high accuracy. In addition, specific cell-cell interactions could be extracted from examples of application to real data in the brain and colon, demonstrating the effectiveness of this method.
→
Journal Publication - Bioinformatics 【DOI】 10.1093/bioinformatics/btac599
CCPLS reveals cell-type-specific spatial dependence of transcriptomes in single cells.
→ Research Outline (PDF In Japanese language)
Digital Medicine Strategy Division - Third Seminar Session
Transborder Medical Research Center
Digital Medicine Strategy Division
Online Zoom Event,
October 12th, 2022 (Wednesday),
18:00 - 19:00
(Please Note: Seminar will be presented in Japanese language only)
→
Digital Medicine Strategy Division - Second Seminar Session
Transborder Medical Research Center
Digital Medicine Strategy Division
Online Zoom Event,
August 30th, 2022 (Tuesday),
18:00 - 19:00
(Please Note: Seminar will be presented in Japanese language only)
→
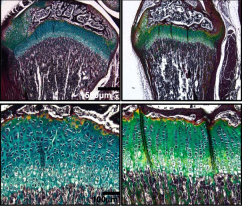
Mast4 determines the cell fate of MSCs for bone and cartilage development.