Development of technology to amplify hematopoietic stem cells in vitro and enable free gene transfer
(Note: Text has been auto translated from Japanese language.)
As a technique for mass amplification of hematopoietic stem cells derived from donor mice in vitro, a method using a medium supplemented with polyvinyl alcohol (PVA) has been reported. Therefore, this research team applied this method to maintain the function of hematopoietic stem cells from bone marrow cells containing a very small amount of hematopoietic stem cells by only immunomagnetic cell separation method using magnetic beads and subsequent simple culture operation. We have developed a technology that can be selectively purified and concentrated as it is. This facilitates gene transfer into hematopoietic stem cells outside the body, and at the same time, it can be amplified in large quantities, making it possible to transplant without using conventional pretreatment such as irradiation. Furthermore, by devising when introducing the target gene, by selecting and transplanting only the hematopoietic stem cells into which the gene has been correctly introduced, the target gene can be expressed in the blood cells in the mouse body at any time after transplantation. Was successful. With this method, there is no tissue damage caused by radiation to the recipient mice after transplantation, and stable breeding and observation can be performed for a long period of time. In addition, there is little invasion and tissue damage to living mice, and it is considered to be an excellent method from the viewpoint of animal welfare. By using these platforms, it has become possible to freely design the function of hematopoietic stem cells according to the purpose of treatment.
→
12, Article number: 3568 (2021)
DOI 10.1038/s41467-021-23763-z
Non-conditioned bone marrow chimeric mouse generation using culture based enrichment
of hematopoietic stem and progenitor cells.
→ Research Outline (PDF In Japanese language)
- Discovery of genes that induce muscle atrophy under microgravity -
(Note: Text has been auto translated from Japanese language.)

In this study, we investigated in detail the effect of gravity on skeletal muscle using a mouse breeding system that can change the gravity environment by a centrifuge developed by the Japan Aerospace Exploration Agency (JAXA). In the Japanese "Kibo" experimental laboratory of the International Space Station (ISS), mice were bred for near on one month in both the micro-gravity environment of space and in an artificial gravity environment (1G) in space. The changes in their muscle was analyzed.
For the first time ever, it has been shown that the artificial gravity environment suppresses the decrease in muscle weight and changes in muscle fiber type and gene expression that occurs in the micro-gravity environment. In addition, we discovered a new gene involved in muscular atrophy that was previously unknown.
The results of this research will provide basic data for long-term manned space exploration on the Moon and Mars, and may also be a key to clarifying part of the mechanism of muscular atrophy seen in the elderly and in the bed-ridden on Earth.
→
doi/10.1038/s41598-021-88392-4
→ Research Outline (PDF In Japanese language)
In order to increase the contribution of donor HSC cells, irradiation and DNA alkylating agents have been commonly used as experimental methods to eliminate HSCs for adult mice. But a technique of HSC deletion for mouse embryo for increase contribution of donor cells has not been published. Here, we established for the first time a procedure for placental HSC transplantation into E11.5 Runx1-deficient mice mated with G1-HRD-Runx1 transgenic mice (Runx1-/-::Tg mice) that have no HSCs in the fetal liver. Following the transplantation of fetal liver cells from mice (allogeneic) or rats (xenogeneic), high donor cell chimerism was observed in Runx1-/-::Tg embryos. Furthermore, chimerism analysis and colony assay data showed that donor fetal liver hematopoietic cells contributed to both white blood cells and red blood cells. Moreover, secondary transplantation into adult recipient mice indicated that the HSCs in rescued Runx1-/-::Tg embryos had normal abilities. These results suggest that mice lacking fetal liver HSCs are a powerful tool for hematopoiesis reconstruction during the embryonic stage and can potentially be used in basic research on HSCs or xenograft models. (Read More –> )
→ Published in Scientific Reports 11, Article number: 4374 (2021)
Key Points
• DHODH inhibition enhances the incorporation of decitabine into DNA in MDS cells.
Environment-based object values learned by local network in the striatum tail

Value and reward are known to be encoded in the part of the brain called the basal ganglia. Research has identified the specific neuronal circuits underlying environment-based value learning.
Obtaining valuables is one of the most important actions for the survival of animals, including humans. And even if they are the same, their value to animals changes depending on the environment and circumstances in which they are placed. Animals make good judgments about their value based on experience and learning. So what is the mechanism by which we learn the value of things?
→ Published in PNAS January 26, 2021 118 (4) e2013623118
Environment-based object values learned by local network in the striatum tail
In skeletal muscle, quiescent satellite cells maintain low levels of protein synthesis, mediated in part through the phosphorylation of eIF2α (P-eIF2α). Paradoxically, P-eIF2α also increases the translation of specific mRNAs, which is mediated by P-eIF2α dependent read through of inhibitory upstream open reading frames (uORFs). Here, we ask whether P-eIF2α dependent mRNA translation enables expansion of satellite cells. We demonstrate that uORFs in the 5’UTR of mRNA for the mitotic spindle stability gene Tacc3 direct P-eIF2α dependent translation. Satellite cells deficient for TACC3 exhibit defects in expansion, self-renewal and regeneration of skeletal muscle.
→ Tsukuba Journal - University of Tsukuba (Japanese language)
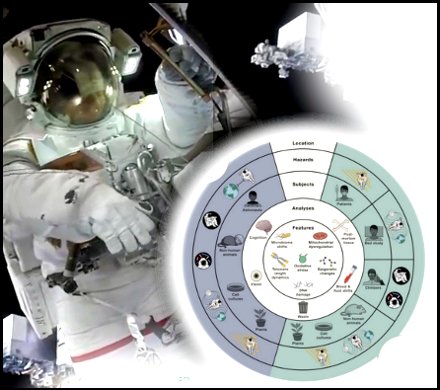
Research on astronaut health and model organisms have revealed six features of spaceflight biology that guide current understanding of fundamental molecular changes that occur during space travel.
→ Cell – Volume 183, Issue 5, 25 November 2020, Pages 1162-1184

In vitro data and molecular dynamics simulations suggest that KS-58 enters cells and blocks intracellular Ras–effector protein interactions. KS-58 selectively binds to K-Ras(G12D) and suppresses the in vitro proliferation of the human lung cancer cell line A427 and the human pancreatic cancer cell line PANC-1, both of which express K-Ras(G12D). Moreover, KS-58 exhibits anti-cancer activity when given as an intravenous injection to mice with subcutaneous or orthotropic PANC-1 cell xenografts. The anti-cancer activity is further improved by combination with gemcitabine.
A New Era for Space Life Science: International Standards for Space Omics Processing
(Published Nov 26, 2020)

TMRC Genomics Division members have been involved in the formation of ISSOP (International Standards for Space Omics Processing).
Scientists across the world are conducting space omics studies to develop countermeasures for safe and effective crewed space missions. However, optimal extraction of scientific insight from such data is contingent on improved standardization.
ISSOP has been founded as an international consortium of scientists who aim to enhance guidelines between space biologists globally.
This paper informs scientists and data scientists from many fields about the challenges and future avenues of space omics and can serve as an introductory reference for new members in the space biology discipline.
→ A New Era for Space Life Science: International Standards for Space Omics Processing
→ Press Release - University of Tsukuba Website (in Japanese language)
→ Research Outline PDF (In Japanese language)
(Published Nov 11, 2020)
Professor Masayuki Matsumoto, TMRC University of Tsukuba, Faculty of Medicine

“Every day of our lives, we make many different decisions. In many cases, we are faced with choosing which of several options would be most rewarding for us. What kind of brain mechanisms might be responsible for the logical decision-making that helps us choose more rewarding behaviors? The key to answering this question is a substance called dopamine, and the neurons that produce it, called dopamine neurons. Prof. Matsumoto is studying the functions of dopamine neurons with the aim of uncovering what role they play in decision-making.”