Molecular Parasitology , RNA Modification and Repair
分子寄生虫学


Molecular Parasitology , RNA Modification and Repair
分子寄生虫学

Department of Infection Biology, Faculty of Medicine
mRNA Cap Formation in Parasitic Protozoan
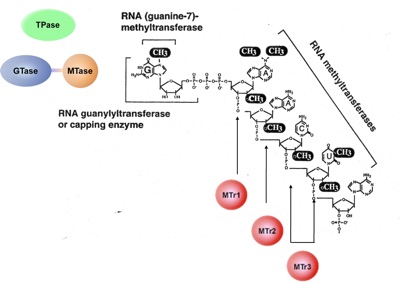
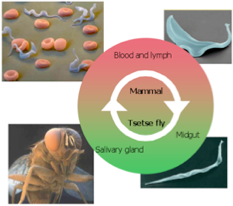
Our primary research interest is to understand the gene expression of eukaryotic parasites with a goal in identifying parasite-specific processes that can be exploited as targets for novel therapeutic interventions. In particular we have focused on how messenger RNA acquire 5’ cap in trypanosomes, parasites that responsible for major public health concerns, such as sleeping sickness in humans and Nagana in cattle. Capping is an essential modification to mRNA to protect the mRNA from degradation and to enhance protein synthesis. We have selected trypanosome as a model system because this organism has a unique form of cap, called cap 4, which consists of the standard cap with additional methylations at seven sites within the first four nucleosides.
Cap 4 is synthesized on the short spliced leader RNA (SL RNA) and then transferred to protein coding pre-mRNA by trans-splicing. Cap 4 and the trans-splicing process are unique to kinetoplastid parasites, which includes Trypoanosoma cruzi and Leishmania spp., which are responsible for Chagas’ disease and Leishmaniasis, respectively. As such, biochemical and molecular characterization of the trypanosome capping enzyme will result in a better understanding of how gene expression is regulated in these parasites and identification of effective drug targets against parasites responsible for a broad range of tropical diseases.
•Characterize the catalytic mechanisms of individual capping enzyme components to determine their unique enzymatic properties.
•Down-regulate expression of the T.brucei capping components by RNAi to examine the consequences of depleting capping enzymes on viability, SL RNA cap synthesis, cap 4 methylation and mRNA trans-splicing in vivo.
•Dissect the mechanism of cap 4 methylation and role of cap 4 in gene regulation in this parasite.
Mechanism of RNA Ligase
Living cells are under constant attack by various environmental and cellular agents that induce breakage of both DNA and RNA. If not repaired, this type of genomic damage can lead to changes in gene expression, mutation, or even cell death. While elaborate DNA repair pathways are understood in reasonable detail, our knowledge of how damage to RNA molecules is recognized and repaired remains very limited. One of the few facts that have been established is that RNA ligase - an enzyme that recognizes breaks in an RNA chain and joins the two ends together - is a key component of this repair process.
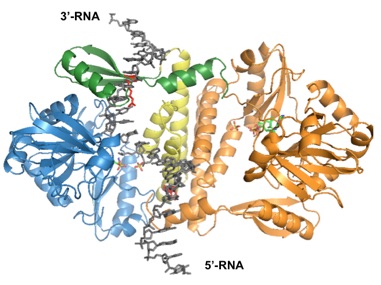
Key questions we aim to address in this project are:
•How does RNA ligase recognize breaks in RNA?
•What are the structural features of RNA ligases that account for its nucleic acid recognition?
•What is the physiological target of RNA ligases?