We are studying evolutionary/adaptation strategies of Gram-positive pathogens. Major research interests include population heterogeneity, and the acquisition of antibiotic resistance. The main research target is the important human pathogen Staphylococcus aureus that inhabits in our nasal cavity but can cause a variety of diseases.
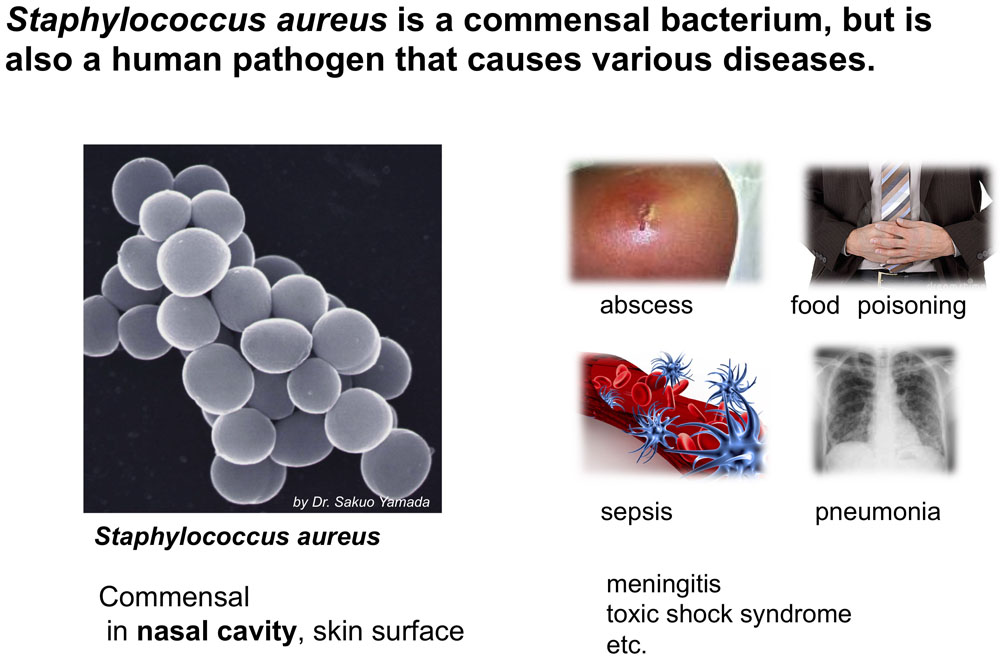
Staphylococcus aureus is a commensal bacterium that naturally inhabits the nasal cavity of mammals, but it is also an opportunistic pathogen responsible for a broad spectrum of infections ranging from food poisoning and superficial skin abscesses to more serious diseases such as pneumonia, meningitis, osteomyelitis, septicemia and toxic shock syndrome. It has acquired resistance to a wide variety of antibiotics, and methicillin-resistant strains (MRSA), the most common cause of nosocomial infections, are now spreading into the community. We are studying evolutionary/adaptation strategies of S. aurus, from our original view points (Microbe Environ,2010).
Natural genetic competence
S. aureus is a major human pathogen responsible for a broad spectrum of infections, emphasized by the emergence of multiple antibiotic-resistant strains with up to 60% of strains worldwide resistant to methicillin (Methicillin Resistant Staphylococcus aureus or MRSA). Many bacteria have the ability to acquire novel genetic characteristics, including antibiotic resistance, through the uptake of extracellular DNA, a phenomenon known as natural genetic transformation or competence. We have shown that the SigH staphylococcal sigma factor is a key component for competence development. Importantly, we have demonstrated for the first time that S. aureus cells producing active SigH become competent for natural transformation, and we were able to confer methicillin resistance to a methicillin-sensitive strain by transformation with chromosomal DNA (PLoS Pathog, 2012). SigH-dependent competence development in S. aureus could help explain the acquisition of antibiotic resistance genes and the rise of the so-called “superbug.” Further research on competence development is necessary to understand evolution of MRSA or upcoming superbugs.
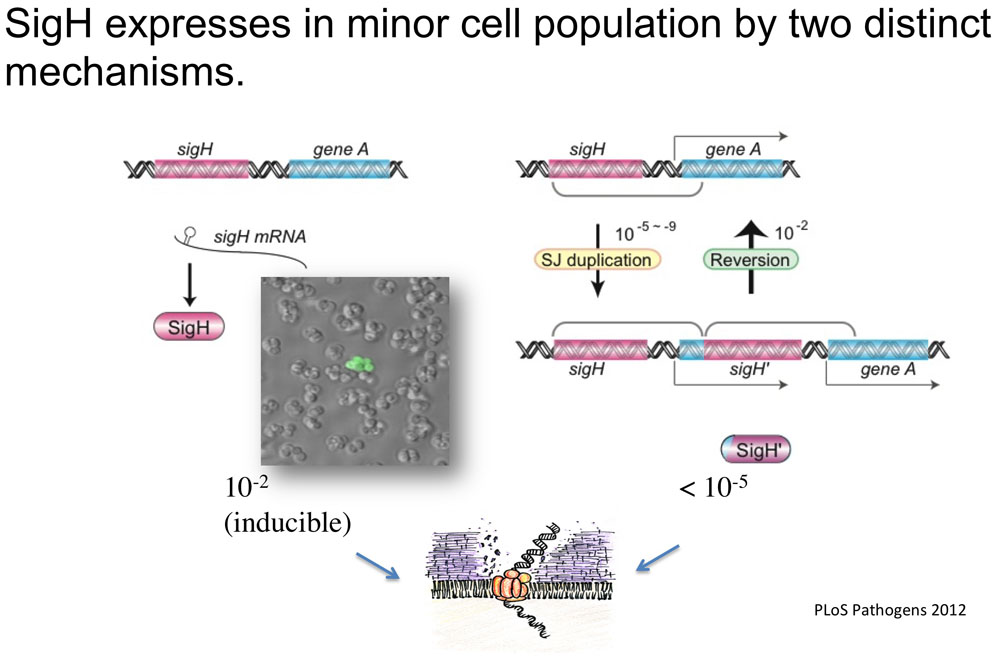
Population heterogeneity
There are growing evidences that the heterogeneity of bacterial cell population plays critical roles in survival, adaptation, and pathogenesis. We found genes that express in minor cell subpopulation (in addition to sigH and competence genes above) and trying to clarify the functions of those genes, aiming to clarify the bacterial characteristics based on the population heterogeneity.
Nucleoid dynamics
In bacterial cells, genomic DNA exists as an architecture termed 'Nucleoid', together with hundreds of nucleoid proteins and nascent RNAs. We are studying the dynamics of nucleoid structure and components in response to environmental stimuli such as oxidative stress and growth phase.
Cardiolipin synthase
Cardiolipin (CL) is an important phospholipid to cope with high salinity or acute acid stress. S. aureus changes the CL content in response to environments by using two CL synthases, one of which (Cls1) specifically works under stress conditions. The stress responsive CL synthesis does not require de novo cls1 expression, but the molecular mechanism is elusive (FEMS Microbiol, 2013).
Adaptation to dryness
S. aureus is naturally quite resistant to dryness: it can survive months on dry surface, which is important characteristics to facilitate the transmission among host. Adaptation to the dryness generates a S. aureus equipped with higher resistance to dryness (Microbes Infect, 2015). This process does not involve the quantitative increase of CL, but CL itself is essential in the acquisition of higher resistance. We have interest in this unknown adaptation mechanism.
Others
In addition, each lab member is studying distinct topics under the scope of 'evolutionary/adaptation strategies of Gram-positive bacteria'
* Interaction of Lactobacillus with other bacteria
* Unique roles of sigH and competence genes in Listeria monocytogenes
* Drug responsive protein Drp35 in Staphylococcus aureus