Transcription factor-binding k-mer analysis clarifies the cell type dependency of binding specificities and cis-regulatory SNPs in humans.
Researchers in the Center for Artificial Intelligence Research and Department of Bioinformatics, Institute of Medicine, at University of Tsukuba, have constructed a new basic data called the “MOCCS profile” of base sequences bound by transcription factors that control human gene expression. Furthermore, they revealed that transcription factors have specific binding sequences for each type of cell and by applying this profile, they established a method to evaluate the effects of genetic variation on DNA binding of transcription factors.
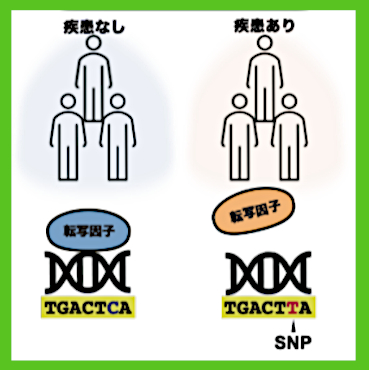
The characteristics of the diverse cells that make up the human body are manifested by differences in gene expression. This control of gene expression is made possible by transcription factors bound to specific base sequences on the genome. It is difficult to clarify the sequences to which transcription factors bind (transcription factor binding sequences) for each cell type, and which are important for elucidating the control mechanism of each gene expression. Until now, the overall picture of transcription factor binding sequences including commonalities and diversity across transcription factor types and cell types had not been clarified.
Researchers used large-scale data on binding sites of human transcription factors to construct new basic data on transcription factor binding sequences, “MOCCS profiles,” and analyzed transcription factor binding sequences across transcription factors and cell types. They conducted analysis of the data and the results revealed that approximately half of the transcription factors analyzed had specific binding sequences for each cell type. Furthermore, by applying MOCCS profiles, researchers developed an index that predicted the influence of single nucleotide polymorphisms (SNPs) on DNA binding of transcription factors. It was shown that it was possible to properly assess the impact.
The MOCCS profile constructed during analysis could be combined with epigenomic data, etc, to help understand cell type-specific gene expression control mechanisms and to evaluate the impact of somatic mutations that occurred in cancer cells on the binding of transcription factors. It is expected that the MOCCS profile can be used in many fields. (Translated from “Tsukuba Journal” - Press Release in Japanese Language - University of Tsukuba Website )
→ Publishing Journal -
BMC Genomics 【DOI】10.1186/s12864-023-09692-9
"Transcription factor-binding k-mer analysis clarifies the cell type dependency of binding specificities and cis-regulatory SNPs in humans."